Abstract
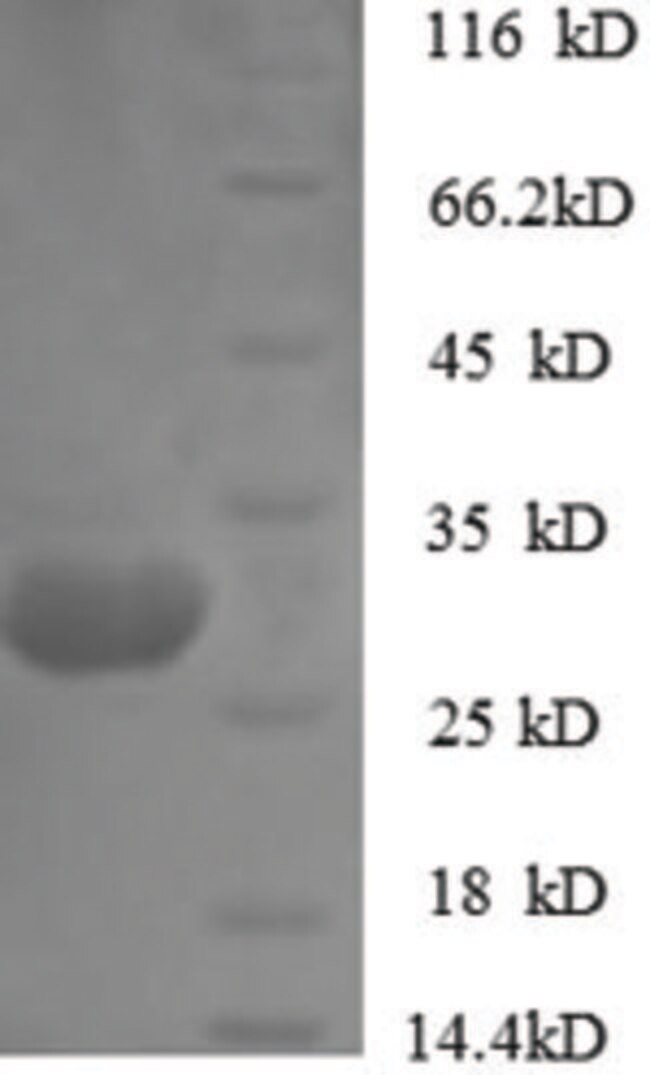
This study was conducted to investigate the expression and enzymatic characteristics of recombinant Oryctolagus cuniculus chymosin (ROCC) expressed in Pichia pastoris. SDS-PAGE of the partially purified supernatant showed two distinct molecular bands of approximately 40 kDa and 45 kDa sizes corresponding to chymosin and partially glycosylated chymosin, respectively. The proteolysis assay showed that rabbit chymosin was more specific compared to bovine and camel chymosins when it comes to hydrolyzing α, β and κ-casein.
Rabbit chymosin maintained its stability over a wide pH range (3.0-6.0) at 37 °C for 8 h. Active chymosin showed maximal enzymatic activity at 40 °C and pH 4.0 with the addition of 75 mM CaCl2. The ROCC coagulation activity in donkey, cow, goat, lamb and camel milk was determined to be 40, 10, 5.7, 3.07 and 2.66 IMCU/mL, respectively. These results revealed that ROCC could have the potential to be incorporated into cheesemaking technology as a milk coagulating enzyme.
Keywords: Cheese; Enzyme; Fermentation; Pichia pastoris; protease; recombinant chymosin.
Extraction of DNA
Total DNA was obtained from the liver or blood by proteinase K digestion, phenol/chloroform extraction and ethanol precipitation using standard techniques. In samples from Las Lomas and Badajoz, purified liver mtDNA was obtained on CsCl gradients (Ennafaa et al., 1987; Biju-Duval et al., 1991).
Amplification
An mtDNA fragment 1120 bp in size, including most of the cytochrome b gene, was amplified with Taq Goldstar according to the manufacturer’s instructions (Eurogentec). The reaction mixture consisted of 50 μL Eurogentec buffer, 150 μM dNTPs, and 1 μM primers. After 3 min of denaturation at 96 °C, PCR was performed in a Kontron cycler for 40 cycles of 1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C. After the last cycle, the PCR was incubated for another 3 min at 72 °C. The primers used were cytb8 (CCCCA TCCAA CATCT CTGCT TGATG AAA; 14242–14269) (Barome et al., 1998) and th3 (GTCTT GTAAG CCAGG AATGG AG; 15340–15361) (Hardy et al., 1994). Numbers indicate nucleotide positions at the 5′ and 3′ ends, respectively, relative to the rabbit mitochondrial genome (Accession number AJ001588, Gissi et al., 1998).
DNA sequence
The sequence of the 1120 bp fragment for an individual carrying the A2Rba haplotype was obtained by direct sequencing using the Sequenase PCR Products (USB) sequencing kit. This sequence was used as a reference in mapping the restriction site of lineage A (Accession number AJ243096).
Data analysis
The GCG MAP SORT subroutine (Devereux, 1989) was used to locate the site gain or loss for each new endonuclease profile. Haplotype and nucleotide diversity and nucleotide divergence were calculated using the REAP package (McElroy et al., 1991). Maximum parsimony was calculated using the heuristic search option of the PAUP software package (Swofford, 1998). Statistical parsimony was used to choose the best network among 100 networks of equal length. PAUP’s ‘show patristic distances’ option was used to identify the closest maximum parsimony network to the actual data (see Templeton et al., 1987, 1992; Templeton & Sing, 1993, for details).